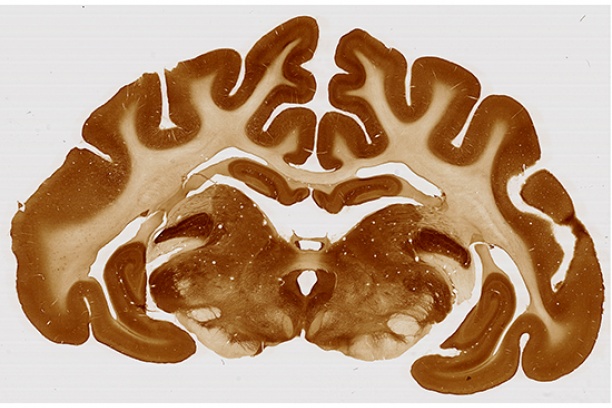
-
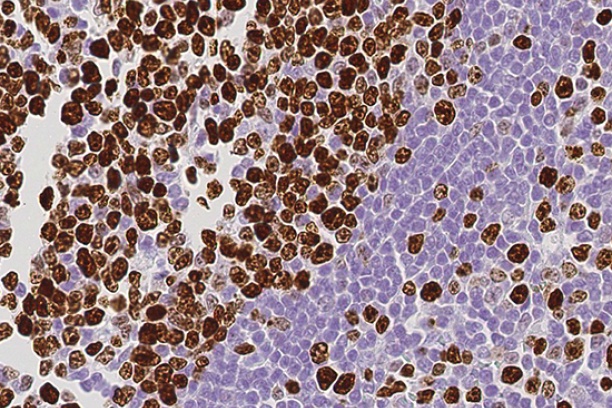
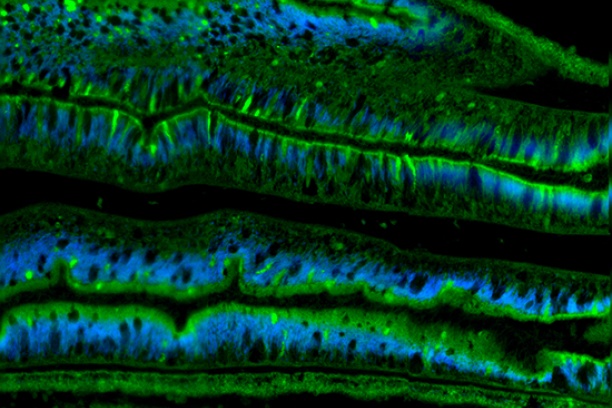
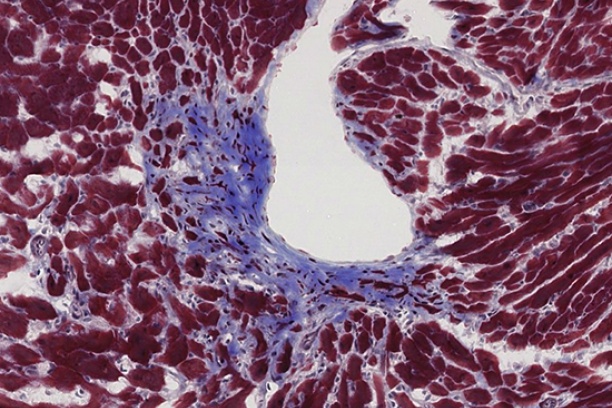
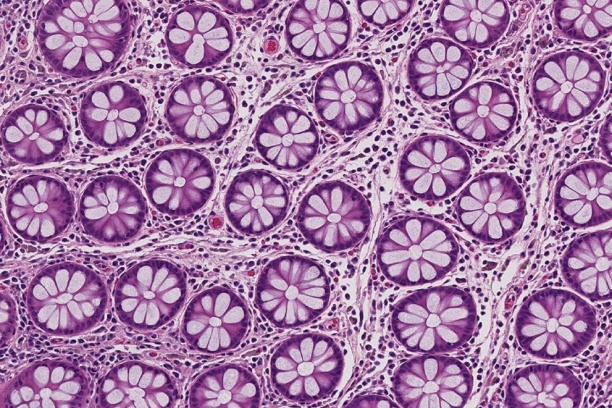
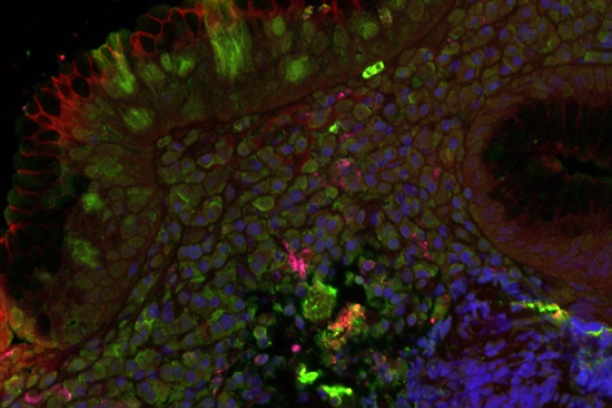
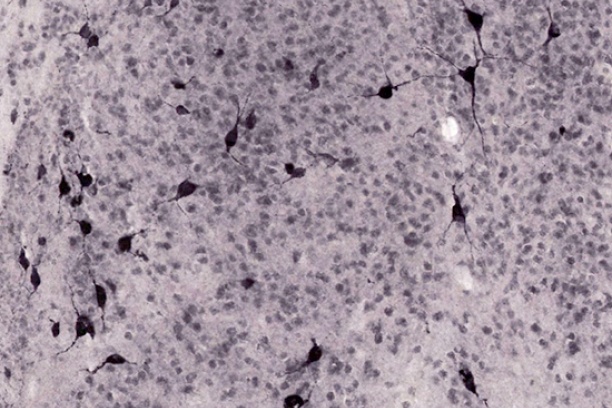
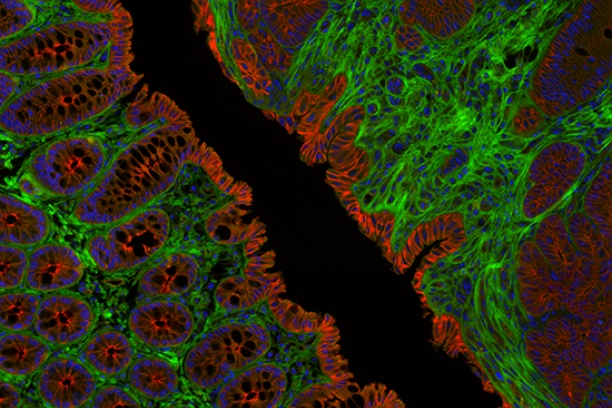
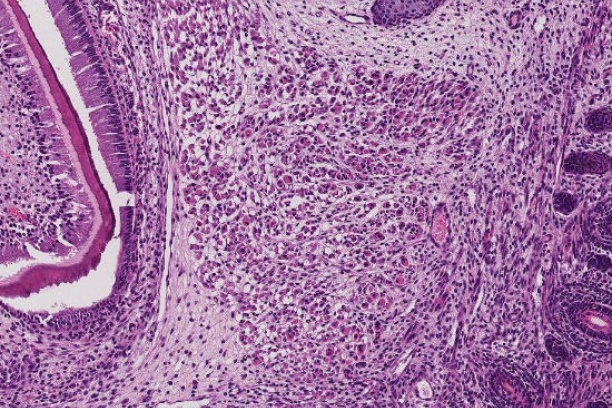
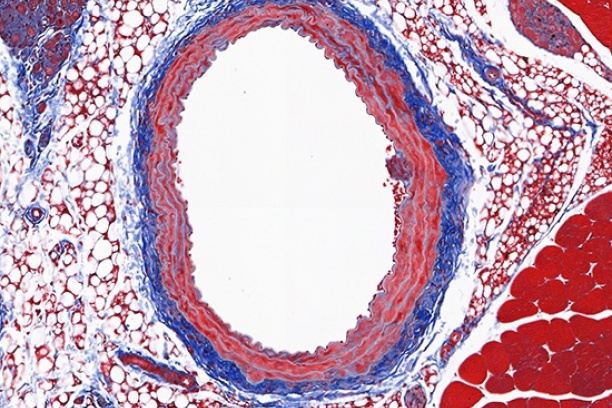
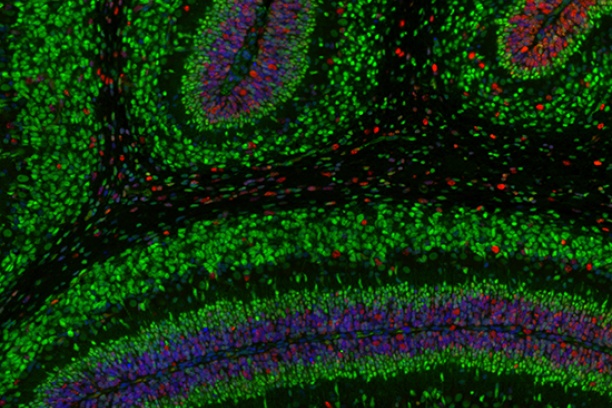
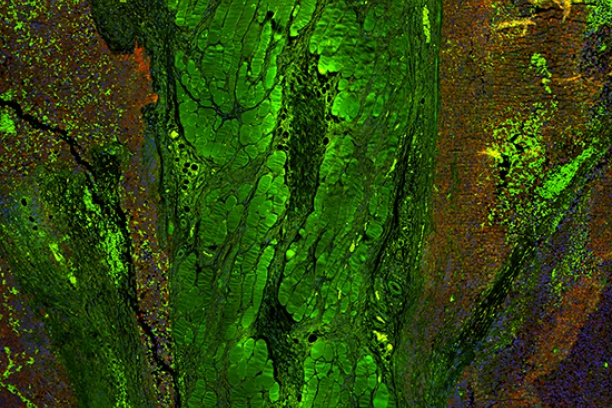
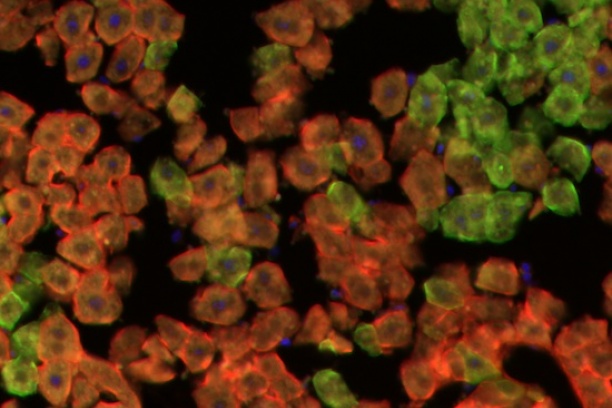
The Digital Histology Shared Resource provides large-scale digital archiving and quantitative analysis of histologic, immunohistochemical and immunofluorescence staining of tissue sections and tissue microarrays. Two Aperio Versa automated scanning microscopes, and Aperio AT2 and a Leica SCN400 Slide Scanner deliver solutions for high-resolution imaging in both bright field and fluorescence. All instruments have high-capacity robotic autoloading (200 slides for the Aperio Versa and Aperio AT2 and 384 slides for the Leica SCN400) making them ideal for large slide cohorts and tissue microarrays. The associated software packages provide complex algorithms for unbiased, automated image analysis and quantitation of immunostaining in both bright field and fluorescence. In addition, these systems can be utilized as a permanent high-resolution solution for those who need archiving of histologic material. The DHSR hosts a convenient, web-based digital slide-viewing environment (Digital Slide Archive) for the rapid retrieval, review, annotation and image acquisition of scanned material. This easy to use digital slide box allows researchers to share images among colleagues from any web browser while remaining in a digitally secure environment. Expert assistance is offered in planning experiments and processing data in a consistent, objective, and timely manner. The automated imaging and analysis performed in this core saves researchers and staff weeks of tedious work. An additional service offered by the DHSR is the creation of digital archives of critical and irreplaceable tissue samples, a benefit only feasible due to the automated high-resolution imaging of whole 25 mm x 75 mm microscope slides and 50 mm x 75 mm "double" slides.
To have slides scanned by DHSR, simply submit a request through iLabs and drop off your slides at 10425 MRB IV.
The DHSR offers specialized microscopy assistance in addition to automated slide scanning and analysis, such as colony counting. The core is capable of high-resolution automated and semi-automated imaging and quantitation of a wide range of cell culture and tissue samples, from bacterial colonies to organoids. The GelCount• system by Oxford Optronix scans and counts mammalian cells, yeast or bacterial colonies in a wide variety of Petri dish and cell culture plate formats. This system is designed for the detection, counting and characterization of stained/adherent mammalian cell colonies or of unstained/non-adherent colonies in soft agar or collagen assays, but also works very well for yeast and bacterial colonies. The software is fully trainable and can be programmed to recognize specific colony features. Detailed information such as diameter, area, density, and nearest neighbor is provided, as well as high-resolution images.
Welcome
Table