
Vanderbilt Genomic Medicine (VGM) Training Program
Program Directors: Josh F. Peterson, MD, Nancy J. Cox, PhD, and Dan M. Roden, MD
The Vanderbilt Genomic Medicine (VGM) Training Program is now accepting applications for the 2025-2026 postdoctoral fellowship. VGM includes a major focus in pharmacogenomics, precision phenotyping, medical informatics, and disease-based genomics.
The program builds on decades-long strengths in critical enabling resources, such as:
- BioVU, the largest biobank linking DNA samples to electronic medical records (EMRs) at a single academic institution (now >244,000 subjects).
- Participation in three NHGRI-funded networks: the Electronic Medical Records and Genomics (eMERGE) network and eMERGE coordinating center, the Implementing Genomics in Practice (IGNITE) Network, and the Undiagnosed Diseases Network (UDN).
- The Pharmacogenomic Resource for Enhanced Decisions In Care and Treatment (PREDICT) project that embeds genetic information in patient EMRs to guide drug and dosage choices (PREDICT served as one model for the ongoing eMERGE-PGx project that implements a similar preemptive genotyping paradigm across eMERGE).
- The Program in Personalized Therapy for Cancer that applies tumor genome sequencing to identify actionable mutations in cancers and to personalized target therapy.
- The HIV-Pharmacogenomics program.
- The largest Department of Biomedical Informatics in the country, with strong research, education, and support programs in clinical information technology.
- Participation and leadership in other related efforts at NIH, including the Genotype-Tissue Expression project (GTEx), the Pharmacogenomics Research Network (PGRN), and the Data and Research Center in the All of Us Program.
The program has 32 highly collaborative and well-funded faculty preceptors who team up to provide both basic and clinical research training opportunities to both PhD-level and MD-level postdoctoral fellows. Trainees will participate in rotations, internships, seminars, journal clubs, and retreats and interact with other trainees and faculty. Under Vanderbilt’s Biomedical Research Education and Training (BRET) Office, the program will provide RCR training, high-standard Individual Development Plans (IDP), and the Career Development programs.
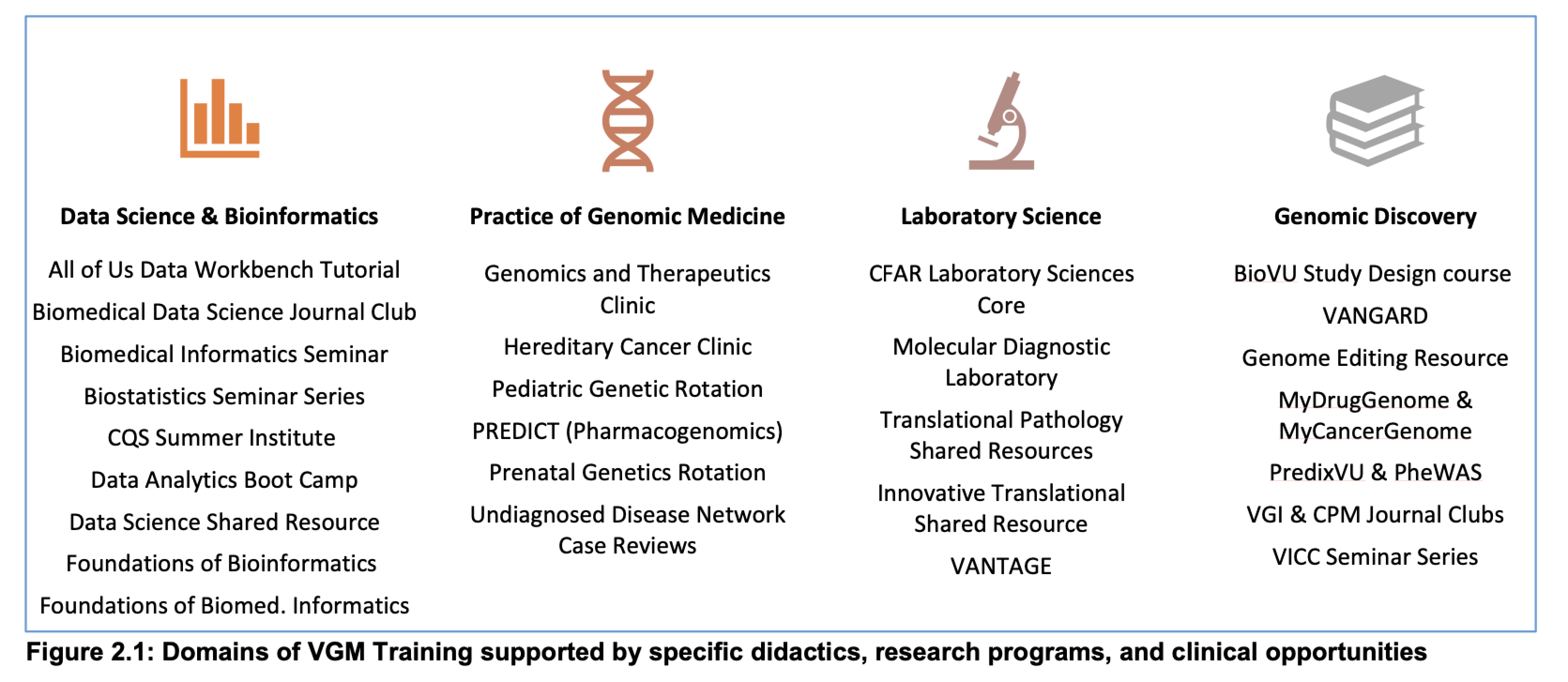
Applicants must have been awarded a doctorate in biomedical sciences, medicine or a related field. Some prior experience in computer science, statistics, informatics, and/or genetics is recommended, but not required.
Anyone interested in being considered must submit the following (in PDF format) to Cynthia Williams:
- 1-page personal research statement
- Letter of recommendation
- CV or NIH biosketch
- Unofficial transcripts
Submissions should reference the program name in the subject line of the email.
If you have any questions about the VGM program, please contact Cynthia Williams, Program Manager, Center for Precision Medicine, Department of Biomedical Informatics.
Acknowledgment: The Vanderbilt Genomic Medicine Training Program is supported by an institutional training grant (T32HG008341) from the National Human Genome Research Institute of the National Institute of Health.
Research Fellow Trainees
-
Mentor:
Dan M. Roden, MD
Professor of Medicine, Pharmacology, and Biomedical Informatics
Director, Oates Institute for Experimental Therapeutics
Senior Vice President for Personalized Medicine
Research Interests:
As more patients are sequenced to identify variants in Mendelian disease genes, there is a growing challenge of interpreting the detected variants. Increasingly, novel “variants of uncertain significance” are being identified, which have little or no published data on their pathogenicity. Variants in the voltage-gated cardiac sodium channel, SCN5A, can lead to Brugada Syndrome and Long QT Syndrome, potentially fatal arrhythmia conditions. The American College of Medical Genetics recommends that incidental pathogenic variants in SCN5A be reported so that patients and family members can be accurately diagnosed and treated. Unfortunately, our lab and others have found that the pathogenicity of SCN5A variants is often unknown or disputed and often does not accurately predict arrhythmias. Identification of a large set of SCN5A variants that have perturbed function in vitro may enable more accurate diagnosis and treatment of arrhythmia syndromes. My aim is to perform a high-throughput in vitro screen of SCN5A coding variants.
I have performed proof-of-principle experiments to demonstrate methods for the four steps of the screen: mutagenesis, transgenesis, functional assays, and high-throughput sequencing. During my time as a T32 fellow, I will apply these methods to survey the channel activity and membrane trafficking of most of the 1920 possible coding variants in an important 96 amino acid region of SCN5A. I will compare the in vitro dataset with known pathogenic variants in this region to determine if the in vitro data can help predict pathogenicity. This work is innovative because it leverages recently developed high-throughput sequencing based methods to broaden and improve our understanding of variants in an important cardiac disease gene. As genomic medicine continues to become more commonplace, the challenge of interpreting patients' variants will continue to grow. This project provides a template for a general approach for improving the breadth and quality of genomic annotations to help deliver on the promise of genomic and precision medicine.
Contact Information:
Email: andrew.m.glazer@vumc.org
Lab: 1285 Medical Research Building IV
-
Mentor:
Jake Hughey, PhD
Assistant Professor of Biomedical Informatics
Research Interests:
Electronic medical records (EHRs) can provide tremendous value in both genomic research and clinical pharmacology to push the frontier of personalized medicine. Recent efforts at Vanderbilt include the development of phenome-wide association studies (PheWAS), which has uncovered new single nucleotide polymorphisms (SNP)-based associations with EHR-derived phenotypes, however SNPs may not always provide the best input variables to stratify patient populations. Additional predictor variables, including more ‘active’ data embedded in e.g. lab tests, narrative data, and self-reported outcomes, may serve as better predictors in a pharmacological setting, such as tracking longitudinal drug efficacy or side effects. Data which may reside outside of typical EHR settings, including social media, and biometric data such as ‘omics profiling, may also provide stronger associations with phenotypes than a more ‘static’ SNP array. Dr. Rhoades aims to develop new predictive tools, incorporating elements of machine learning and natural language processing, to increase the capacity of PheWAS analyses for clinical decision-making.
Contact Information:
Email: seth.d.rhoades@vanderbilt.edu
Lab: 2525 West End Ave, Suite 1500
-
Mentor:
Nancy J. Cox, PhD
Director, Vanderbilt Genetics Institute
Professor of Medicine, Division of Genetic Medicine
Director, Division of Genetic Medicine
Mary Phillips Edmonds Gray Professor of Genetics
Research Interests:
As a trained cellular biologist and neuroscientist, I hope to expand my current knowledge of genetics and biological processes to better understand the relationships between genetic sequences and human disease. For the past 8 years, I have examined basic biological problems, first in cancer cells as a research technician, followed by my graduate work with neurons. I studied cancer progression and neuroplasticity at the cellular level, utilizing genetic and biochemical tools to dissect complex molecular pathways. Therefore, I feel that my expertise in investigating basic biological functions will allow me to approach genomic science with a unique perspective.
I joined David Miller’s lab as a graduate student in 2011, where I worked to uncover a genetic pathway that stabilizes synaptic connections in the nematode C. elegans. Using genetic tools, I found that a conserved sodium channel protein, UNC-8 (a degenerin/epithelial sodium channel protein) drives synapse removal in a calcium-dependent pathway. The phosphatase calcineurin also promotes synapse disassembly in this pathway, which is regulated by both transcriptional regulation and neuronal activity. This research aided in my training in several genetic and molecular techniques, including: live-imaging, immunofluorescence, image analysis, molecular biology, genetics, optogenetics, behavioral assays, CRISPR technology, biochemistry, and statistical analysis.
In addition to my scientific training, as a graduate student I developed strong written and oral communication skills through writing grant proposals, authoring manuscripts, and presenting my data at regional and international conferences. I mentored several undergraduate, graduate, and medical students in the lab and served as a teaching assistant in a graduate level course to strengthen my mentoring skills. Collaboration was also critical for my success as a graduate student. I organized monthly Skype meetings with two collaborating labs. These meetings were crucial to the publication of the eLife manuscript in which I am first author, and allowed me to hone my communication, organizational, and project management skills. These experiences helped me to develop transferable skills that will be essential for my success in all research settings.
As a postdoctoral fellow, I plan to build upon my bench skills by using computational and quantitative analysis of the genome to better understand human diseases. I am fascinated by the complexity of the genome and how sequence aberrations lead to disease. The Vanderbilt Genomic Medicine Postdoctoral Fellowship will provide an opportunity for me to utilize the modern tools that have been developed at Vanderbilt, including the BioVU genomic sequence bank and the pheWAS catalog. By combining the tools and training available in the VGM program and my strong background in molecular genetics and biology, I can develop a deeper understanding of the genome and how sequence changes result in phenotypic outcome. This information can then be applied to improve the diagnosis and treatment of human patients.
Contact Information:
Email: tyne.w.miller-fleming@vumc.org
Lab: 507 Light Hall
-
Mentor:
Todd Edwards, PhD
Assistant Professor of Medicine, Division of Epidemiology
Associate Director, Vanderbilt Genetics Institute
Research Interests:
My methodological research addresses the challenges of understanding the genomic architecture of complex human traits, while my applied research extends this to discover the genes and genomic mechanisms underlying traits in specific populations. An innovative aspect of my work is that genes associated with the physiological traits related to a disease may be involved in epistatic interactions with novel variants. This perspective has led to the development of a powerful and cost-effective methodology to detect genetic associations that otherwise would require much larger studies to observe marginal effects. The motivation for this work is the well-established missing heritability problem, and the intention to detect genetic determinants missed by univariate association scans. I have contributed to this area of research by modeling first-order interactions with genetic variants, or multiple variants summarized in a genetic risk score, that are associated with physiological traits closely-related to a disease, measured by high-quality, dynamic phenotypes in small cohorts, to detect associations with the trait of interest in larger case-control cohorts. This approach alleviates the high computational costs and multiple-testing burden that has hindered previous gene-gene interaction investigations.
I developed this methodology in response to the observation that genome-wide investigations of type 2 diabetes (T2D), a disease that affects hundreds of millions of individuals globally, has led to the discovery of variants that only explain 10-30% of disease heritability. Further, the majority of my work has been centered on cardiometabolic disease in African Americans, a classically understudied population that has a high burden of disease. The most recent data indicates that the prevalence of T2D among African Americans (13.2%) is much higher than that of European Americans (7.6%). The application of my methodological work is a powerful collaborative consortium project using resources from the ARIC, CARDIA, JHS, and MESA cohorts examining gene-gene interactions contributing to T2D risk in African Americans.
In addition to my work analyzing gene-gene interactions, I have also worked on other collaborative projects dealing with many types of data. I have led meta-analyses in the MEDIA project, an African American T2D genome-wide association study (GWAS) meta-analysis incorporating over 30,000 individuals from approximately 20 studies, alongside Dr. Maggie Ng from the Center for Diabetes Research at Wake Forest. This has led to the development of cutting-edge analytical approaches and highly effective quality control procedures, and has allowed for the inclusion of data from this project in the DIAMANTE consortium, a trans- ethnic T2D meta-analysis incorporating over 700,000 individuals from 5 different ethnicities.
Also, I have developed a technique that allows for simultaneous modeling of association, two point linkage, linkage conditioned on association, association conditioned on linkage, and joint linkage and association in family-based studies in an effort to identify high-impact, putatively causal variants. We are applying this approach to whole-exome microarray data from African American, Mexican American, and European American individuals from the IRASFS and DHS studies in an investigation of the genetic determinants of lipid metabolism traits. My research in various racial and ethnic groups, a challenging yet rewarding experience, has contributed to the understanding that only a portion of the genomic architecture of cardiometabolic traits are shared across ethnicities and has highlighted the need for further genetic investigations in multiple ancestries.
My future goals as a researcher are to obtain a position as a tenure-track faculty member at a major research center and conduct association studies in electronic medical record databases, such as the eMERGE network and BioVU at Vanderbilt, the Million Veteran’s Project, or the All of Us initiative. I would further like to pursue the development of statistical and computational methods for high-throughput omics (i.e. transcriptomics, metabolomics, etc.) data integration to identify targets for downstream molecular, cellular, and organismal-level functional investigations of candidate disease loci. Considering my research experience, my background in biochemistry and genetics, and my extensive training in computational methods, I believe I am a good candidate for this funding opportunity. I believe the Edwards lab at Vanderbilt is an ideal environment for the continuation of my research and training, as they are already involved in research activity in my target domains. I also feel that the Vanderbilt Genetics Institute is a dynamic and rapidly growing enterprise with access to outstanding resources and opportunities for trainees to develop into independent academic scientists.
Contact Information:
Email: jacob.keaton@vumc.org
Lab: 2525 West End Ave, Suite 800
-
Mentor:
Jeremy L. Warner, MD, MS
Associate Professor of Medicine and Biomedical Informatics
Medical Director, Vanderbilt Cancer Registry & SCT Data Analysis Team
Research Interests:
I am preparing for a career in malignant hematology, with a focus on plasma cell disorders (multiple myeloma and AL amyloidosis). These disorders are not curable but are highly treatable with many novel therapeutic agents having been approved for routine use in recent years. Given the chronic nature of the diseases and potential for delayed effects of treatment, a critical question in the field is how to identify patients that are likely to experience toxicity of therapy. In particular, many of the agents that are used to treat plasma cell diseases are associated with peripheral neuropathy, which can be a debilitating complication. I am currently contributing to ongoing efforts to use BioVU to identify patients at risk for chemotherapy-induced peripheral neuropathy from microtubule inhibitors. Although these are not used to treat plasma cell diseases, I hope to learn skills which I can apply to similar analyses of neuropathy associated with different agents.
Contact Information:
Email: samuel.m.rubinstein@vumc.org
Lab: 2525 West End Ave, Suite 1500
-
Mentor:
Travis J. Osterman, D.O.
Assistant Professor of Medicine and Biomedical Informatics
Research Interests:
A large portion of pathology relies upon making sense of visual information, which is increasingly becoming digital. With the arrival of increased computing speed, rapid networking, cheap storage, and advancements in probabilistic modeling advancements, digital pathology applications have become increasingly utilized in pathology practices.
Digital pathology is the practice and science of turning digitized histopathological images from whole slide scanning into quantitative actionable information. It can be used for research and the development of clinical decision support systems such as rapid retrieval of prior cases, remote collaboration for interpretation of diagnoses, EMR integration, and image analysis.
The application of digital pathology image analysis via neural networks quantification of histologic and cytologic features, and tissue biomarkers allows for greater accuracy and reproducibility than human-based assessment alone. It will also enable the capture of subvisual morphometric phenotypes. This will enhance the scope and performance of precision medicine that aims at developing patient-tailored therapies.
The aim of my research is to configure deep learning neural networks for image analysis that facilitate diagnosis, grading, prognostication, prediction of the response to therapy and the creation of large well-curated validation datasets. Other research interests include the development of end-to-end digital lab work including scanning, slide sharing, and annotation as well as the development of clinical decision support systems for both anatomical and clinical pathology.
Contact Information:
Email: peter.c.louis@vumc.org
Lab: 2525 West End Ave, Suite 1500
-
Mentor:
Digna Velez-Edwards, Ph.D.
Associate Professor of Ob/GYN
Research Interests:
The Developmental Origins of Health and Disease theory suggests that an individual’s health, from infancy through adulthood, is significantly influenced by exposures during the gestational and neonatal periods. A growing body of epidemiologic research supports this theory, identifying the perinatal period as a critical time for overall lifelong health of mothers and their children. My research builds off of this theory by focusing on the perinatal period. My research examines women’s health prior to and during pregnancy and the long-term health consequences of exposures in these periods for both the women themselves and their children. Leveraging large electronic medical record databases linked to genomic data, I also hope to identify early life epidemiological and biological factors related to health outcomes in infancy and early childhood. The overall goal of this line of research is to identify and understand epidemiologic and genetic risk factors that impact the life-long health of women and children.
Contact Information:
Email: elizabeth.jasper@vumc.org
Lab: 2525 West End Ave, Suite 1500
-
Mentor:
Raymond Blind, Ph.D.
Assistant Professor of Medicine, Biochemistry & Pharmacology
Division of Diabetes, Endocrinology & Metabolism
Research Interests:
Nuclear receptors are a superfamily of 48 transcription factors, each with various confirmed patient polymorphisms that correlate with a wide array of human diseases. All 48 nuclear receptor genes encode proteins that share a similar domain structure, composed of a DNA-binding domain connected to a lipid-binding transcriptional-activation domain. The structural biology describing these two individual domains has explained many disease-relevant polymorphisms, but structural models of the isolated domains have failed to explain many others. To address this, the Blind lab recently developed a computational structural modelling pipeline, used to describe how the interface between the two domains of the nuclear receptor NR5A2 are structured. These data show that a human polymorphism in the coding sequence from the gnomAD database (S148R) exists in this interface, with wet lab data showing how the polymorphism alters the structure and function of the NR5A2 protein. Taking a similar computational modeling approach, I am developing a model of how the two domains of the NR5A1 nuclear receptor interact, intended to be used in precision medicine efforts to determine how and why particular polymorphisms in NR5A1 result in loss-of-function and human diseases, such as 46 X,Y sex reversal and adrenal insufficiency. I am also examining how polymorphisms affect ligand activation of NR5A1 and NR5A2 using in silico docking. Together, my work will elucidate how long-unexplained disease-associated polymorphisms in human patients alter NR5A structure.
Contact Information:
Email: zeinab.haratipour@vumc.org
Lab: 2525 West End Ave, Suite 1500
-
Mentor:
Dan M. Roden, M.D.
Professor of Medicine, Pharmacology, and Biomedical Informatics
Director, Oates Institute for Experimental Therapeutics
Senior Vice President for Personalized Medicine
Research Interests:
I am interested in using polygenic methods to improve clinical classification and treatment strategies. For example, the risk of developing type 2 diabetes and many other common diseases, is influenced by polygenic risk, which is the cumulative, small effects of many common gene variants. A number of studies have also suggested that variability in polygenic disease risk also affects variability in response to drug therapy for common disease. Polygenic risk can be quantified for an individual using polygenic risk scores (PRS). Recent studies in coronary artery disease and drug-induced long QT syndrome have shown the PRS developed for a disease or phenotype are also able to predict response to drugs in treatment of that disease. We plan to apply that idea to treatment of type 2 diabetes. If there is an interaction between polygenic risk and treatment response, this may identify patients who will benefit more or less from specific treatments, and inform clinical practice.
Contact Information:
Email: megan.lancaster@vumc.org
-
Mentors:
Jonathan Mosley, M.D., Ph.D.
Assistant Professor
Division of Clinical Pharmacology
Departments of Internal Medicine and Biomedical Informatics
Jane Ferguson, Ph.D.
Assistant Professor
Division of Cardiovascular Medicine
Department of Medicine
Research Interests:
I am interested in epidemiological, multi-omic translational research in nutritional sciences. I believe that translational research in genomics and metabolomics represents a promising avenue to contribute to the evidence base supporting precision nutrition approaches and can reveal the mechanisms of the relationship between environmental factors (such as physical activity and diet) and diseases. Integrating multi-omics data with clinical and environmental data could also improve our understanding of the molecular basis of disease, prediction of therapeutic responses and risk of adverse effects, and implementation of new tools towards the goal of more personalized medicine.
Contact Information:
Email: minoo.bagheri@vumc.org
-
Mentors:
Alexander Bick, M.D., Ph.D.
Assistant Professor
Division of Genetic Medicine
Department of Medicine
Dan M. Roden, M.D.
Professor of Medicine, Pharmacology, and Biomedical Informatics
Director, Oates Institute for Experimental Therapeutics
Senior Vice President for Personalized Medicine
Research Interests:
Advances in population scale genomic sequencing have greatly expanded the understanding of the inherited basis of cardiovascular disease. Reanalysis of these genomic datasets identified an unexpected risk factor for cardiovascular disease (CVD), somatically acquired DNA mutations. Clonal Hematopoiesis of Indeterminate Potential (CHIP) represents the most commonly identified form of somatic mosaicism and drives poor outcomes across a spectrum of CVD. Though these mutations are commonly found in those older than 60, little is known about how these mutations exert a pathological influence in the setting of cardiovascular disease. Combined with cutting edge bulk sequencing, emerging but low-throughput methods such as single cell DNA sequencing and multi-omics we hope to shed light on the accumulation of mutations throughout the life of a particular cell and reveal critical aspects of pathophysiology as it pertains to cellular function over time. These insights will open the door to new therapeutic modalities and the treatment of CVD.
-
Mentor:
Lea K. Davis, Ph.D.
Associate Professor of Medicine
Department of Medicine, Division of Genetic Medicine
Associate Professor of Biomedical Informatics
Associate Professor of Psychiatry and Behavioral Sciences
Research Interests:
As a molecular and genetic epidemiologist, I research how environmental and social risk factors interact with genetic factors across the life course. I use techniques from epidemiology, biostatistics, and bioinformatics to leverage genomic data in observational cohorts, with an emphasis on the creation of biomarkers. By developing biomarkers which explicitly - acknowledge or incorporate environmental and social factors, my work helps to identify susceptible populations, develop targeted interventions, and address health disparities.
One important sociobiological contact which modifies biomarker performance and disease risk is sex. Yet many clinical biomarkers are based on small samples of men, even while distributions of clinical laboratory values differ by sex. My work explores how sex modifies genetic determinants of phenotypes, and subsequent disease incidence and progression. Understanding sex-differences in clinical biomarkers can help identify and alleviate disparities in health care. Sex differences in the clinical phenome are also suggestive of important differences in immunity and inflammatory responses. I'm particularly interested in understanding how genetic factors might interact with biotic exposures - including infectious disease and the microbiome - to impact chronic health conditions, including neurological and psychiatric conditions.
-
Mentor:
Brian R. Lindman, M.D., M.S.C.I., F.A.C.C.
Associate Professor of Medicine
Medical Director, Structural Heart and Valve Center
Vanderbilt University Medical Center
Research Interests:
I am focused on identifying whether proteomic signatures of myocardial remodeling/dysfunction are present early in aortic stenosis before traditional clinical thresholds for intervention. If present, this may point to known/novel pathways contributing to heart failure in aortic stenosis patients and provide an early molecular barometer for clinical surveillance and personalized timing for aortic valve replacement.
-
Mentors:
Douglas Ruderfer, Ph.D.
Associate Professor of Medicine (Division of Genetic Medicine)
Associate Professor of Psychiatry and Biomedical Informatics
Vanderbilt University Medical Center
Lisa Bastarache, M.S.
Research Associate Professor
Department of Biomedical Informatics
Scientific Director for PheWAS Core
Vanderbilt University Medical Center
Research Interests:
Broadly, I am interested in the biological impact of rare genetic variation on complex and Mendelian diseases. Using a variety of statistical and bioinformatic techniques, large whole genome sequenced (WGS) cohorts with paired data from the electronic health record (EHR) present an opportunity to investigate genotype-phenotype associations. As part of an effort with the Vanderbilt Undiagnosed Diseases Program, I am developing a method that identifies the similarities of their clinical presentation to support diagnoses utilizing WGS. In addition, I am interested in techniques that inform the interpretation of the overwhelming number of variants of uncertain significance that hinder the ability of clinical genetic sequencing to return meaningful results for patients and providers. By leveraging a biologically diverse sequenced cohort, I aim to determine the relationship between established pathogenic variants and their observed impact on a patient's phenotype and compare outcomes in patients carrying VUS in the same genes.
-
Mentor:
Piper Below, Ph.D.
Professor of Medicine
Division of Genetic Medicine
Robert A. Goodwin Jr., M.D., Directorship in Medicine
Vanderbilt University Medical Center
Research Interests:
I am a Vanderbilt Genomic Medicine (VGM) postdoctoral fellow and a research fellow in the Below Lab with broad interests in data science and biomedical informatics, relating to applying both novel and extant methods toward building models that predict a wide variety of outcomes. I joined the Below Lab in 2023 and the VGM Training Program in 2025. My current research focuses on developing machine learning and statistical methods for multi-omics approaches to gene discovery for a wide variety of phenotypes. My long-term goal is to develop a research program integrating electronic health records, and multi-omics data to develop clinical outcome prediction models that encompass the broadest possible array of factors that contribute to health.
Predoctoral Research Training PhD in Biomedical Informatics
MD-PhD Program (MSTP)Postdoctoral Research Training NLM Postdoctoral Fellowship in Biomedical Informatics
VA Postdoctoral Fellowship in Medical Informatics and Quality Improvement
Vanderbilt Genomic Medicine Fellowship
Additional postdoc position listingsApplied Clinical Informatics Training MS in Applied Clinical Informatics
Clinical Informatics Fellowship
Summer Research InternshipBiomedical Informatics Summer ProgramContinuing Professional EducationDBMI Grand Rounds
Vanderbilt Clinical Informatics Practical Shorts (VCLIPS)