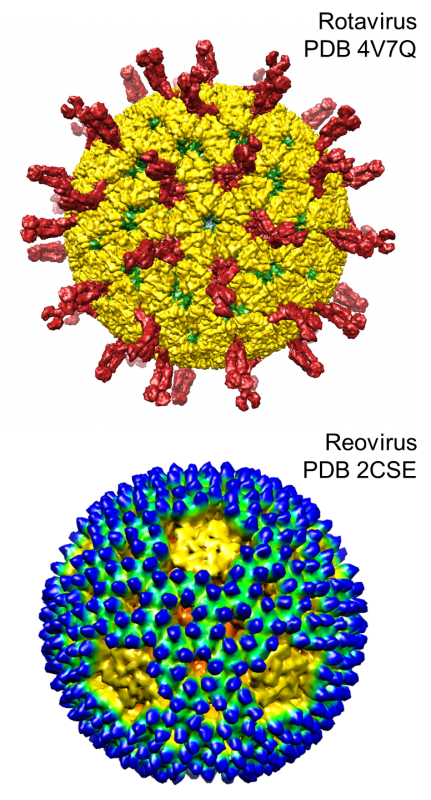
RNA viruses are ubiquitous intracellular parasites that cause a number of important diseases. In order to successfully replicate in the different tissues and hosts they encounter, viruses must be able to efficiently bind, enter, and replicate in target cells and to adapt rapidly to environmental pressures. Several features, including large population size, fast replication rate, and short generation time, permit populations of RNA viruses to evolve much more rapidly than DNA-based organisms. These features also enable us to study viral evolution in real time. Research in the Ogden lab is focused on the tropism and diversity of segmented, double stranded RNA viruses of the order Reovirales, including rotavirus and mammalian orthoreovirus (reovirus).
Rotavirus is an important cause of diarrheal disease that results in the deaths of hundreds of thousands of infants and young children worldwide each year. Licensed vaccines have significantly reduced the burden of rotavirus disease in several countries, but they are generally less effective in high-mortality. low- and middle-income countries than low-mortality, high-income countries. Currently, our lab is studying how rotavirus outer capsid proteins contribute to the responses we generate against vaccines and mediate the cell-type specificity of infection. We are also investigating the functions of nonstructural proteins in viral replication and spread for less well studied rotavirus species.
To infect and adapt, viruses in the order Reovirales have evolved mechanisms to promote viral genetic diversity. Evolutionary benefits of diversity are countered by a need to maintain interactions mediating packaging of a multi-segmented genome, assembly, egress, and transmission. Thus, the replication strategy also may inherently constrain diversity. Our laboratory uses reovirus as an experimental system to study how dsRNA virus assembly and transmission processes regulate unique aspects of viral genetic diversity. Some of the themes we are investigating include the identification of RNA sequences required to package the multi-segmented genome, generation and packaging of defective viral genome segments, genome segment reassortment during coinfection and superinfection, mechanisms of reovirus egress, and effects of multi-particle infection.