Juan Zhao, QiPing Feng, Patrick Wu, Roxana A. Lupu, Russell A. Wilke, Quinn S. Wells, Joshua C. Denny & Wei-Qi Wei*
We developed machine learning and deep learning models to predict 10-year CVD risk using longitudinal EHR and genetic data.
Cardiovascular disease (CVD) is the leading cause of morbidity and mortality worldwide. Existing models such as Framingham risk score, ACC /AHA pooled equations, are based on a small number of risk factors such as hypertension, cholesterol, age, smoking, and diabetes, which can't capture people with few risk factors. In this project, we examined whether using machine learning (ML) and deep learning (DL) on longitudinal EHR data and incorporating genetic information can improve CVD prediction.
|
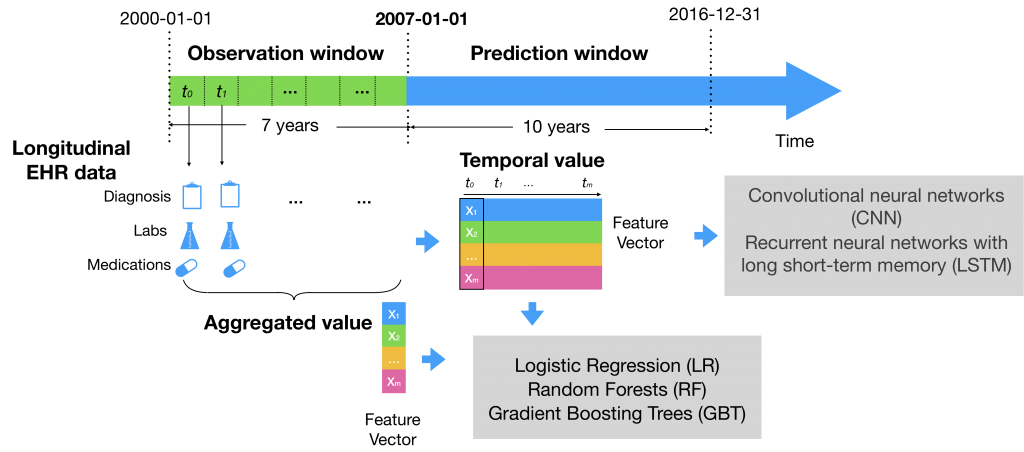
Besides conventional risk factors, we included more variables in EHRs data such as vital signs, lab tests, previous diagnosis code, and medication history. We compared two ways of modeling the data: a) using aggregate values (mean, median, max, min, and standard deviations) of each numerical values and b) using temporal series values prior to developing a CVD. We applied logistic regression, GBT, and LSTM. |
 |
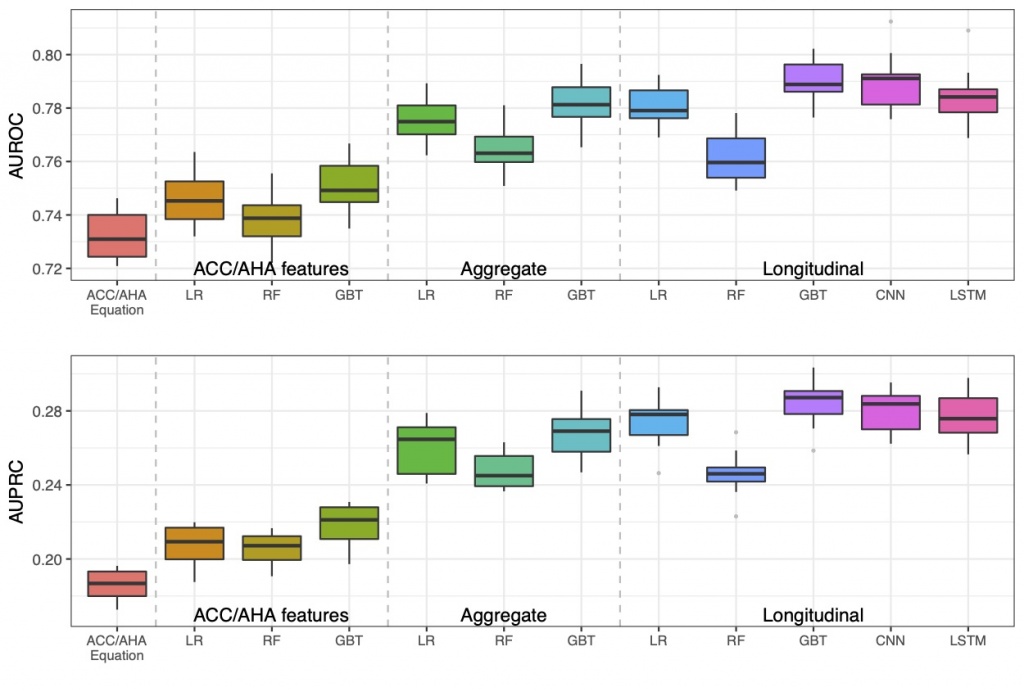 |
Using longitudinal features outperformed using aggregate features. We used a large dataset of EHR at Vanderbilt University Medical Center, which included 109, 490 adults. We trained the models using nested 10-fold cross-validation. The best models were gradient boosting trees (GBT) and LSTM with temporal/longitudinal values. The best AUROC was 0.79, significantly outperformed the AUROC of 0.73 achieved by the baseline approach – AHA Pooled Cohort Risk equations. |
|
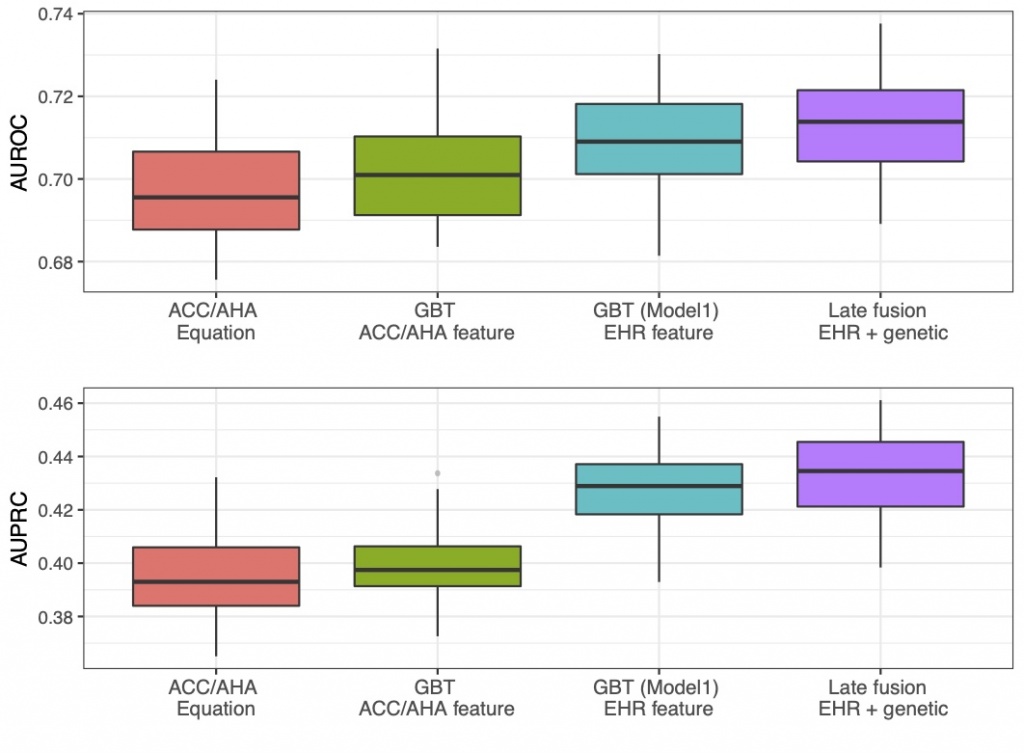
Adding genetic data using a late-fusion model improved the prediction performance. We incorporating genetic data into the feature set. Genetic data could provide an important role in explaining many CVD events without traditional clinical risk factors. But previously, how to leverage genetic information for chronic disease prediction remained a challenge. We notably developed a late-fusion model to combine the genetic and clinical together. Our models have achieved better performance compared to the baseline approach (AUROC of 0.713 vs. 0.698). |
 |
To learn more, read our full paper, or get the source code.
If you have questions about our work, contact us at: juan.zhao@vumc.org and wei-qi.wei@vumc.org